We recently reported our work constructing single-heme proteins from our water-soluble and transmembrane diheme proteins 4D2 and CytbX. Whilst we initially set out to make these proteins for ‘completionist’ reasons, we realised we could use them to gather valuable insights to the factors that lead to redox potential splitting in our diheme proteins.
Read the article here: https://onlinelibrary.wiley.com/doi/10.1002/pro.5113
“With the successful 4D2 and CytbX designs in hand, we are in the unique position to systematically study the interactions between a pair of b-type hemes within the same soluble protein scaffold, and directly compare them with the membrane equivalent.”
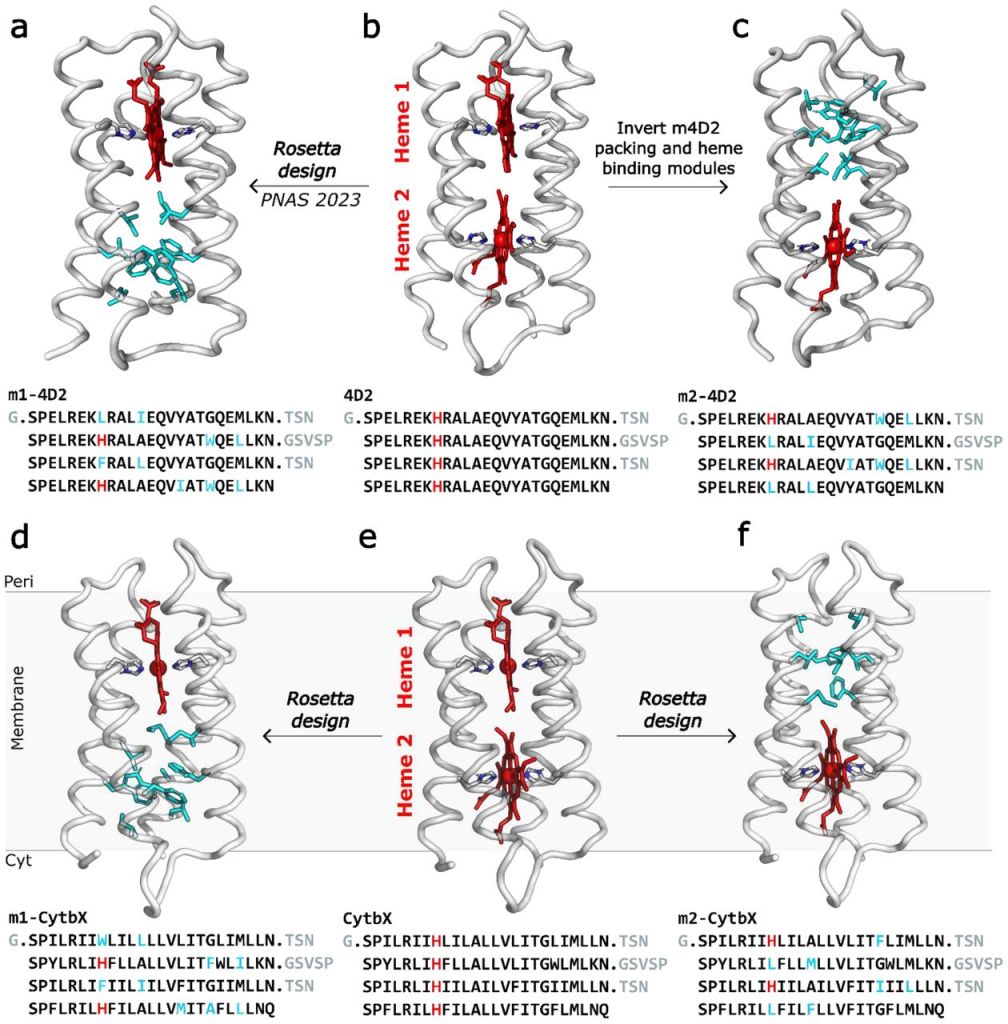
By comparing the redox potentials of the water-soluble and transmembrane single-heme and di-heme proteins, both experimentally and computationally, we deduced that individual binding sites within the protein have minimal impact on the redox potential split, with heme-heme electrostatic interactions being the major contributor.
Additionally, by combining results from two different computational continuum-electrostatics methods (PB-MC and BioDC), we observed that burial of the heme pair greatly strengethens the magnitude of heme-heme electrostatic coupling, leading to the greater split of redox potentials in CytbX compared to 4D2.
We believe this likely extends to natural multiheme membrane cytochromes, partly explaining the large split of redox potentials (around 100 mV) commonly observed in these proteins.
This work also speaks to the robustness of our core designs and scaffolds. We can make large-scale changes to each half of the protein and obtain constructs that are still highly stable and readily expressible in E. coli.
Leave a comment